Phylogeny
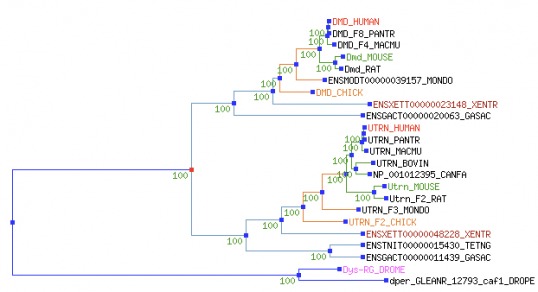
For phylogenetic analysis of dystrophin, the protein sequence was used due to the extremely large size of the dystrophin DNA sequence. Using protein sequence for phylogenetic tree creation also excludes untranslated regions, including introns, which may be less evolutionarily conserved than the protein (or mature mRNA sequence). Treefam was exclusively used because of protein-sequence size restrictions present in other phylogenetic tree generating sites.
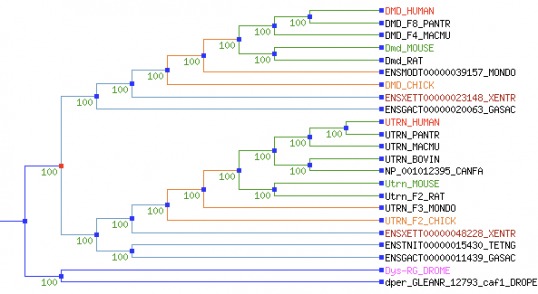
Both phylogenetic trees above used identical algorithms to calculate similarity of the dystrophin protein across species. The second figure is adjusted so that the different tree branches are not proportional to variation in protein sequence (as in the first figure) to allow for more easy visualization of the tree. It is notable that all branches labelled with "Utrn" refer to a very similar protein to dystrophin called utrophin. Clicking on the top tree will bring up a larger image, while clicking on the bottom tree will link to the original site of data retrieval. This is a very interactive website, and it is highly recommended for those who want to see full descriptions of the phylogenetic relationships.
References
"Human DMD Phylogenetic Tree." TreeFam. Web. 5 Mar. 2010. <http://www.treefam.org/cgi-bin/TFinfo.pl?ac=TF320178>.
This site was created by Anthony Parendo for an undergraduate Genetics 677 course at the University of Wisconsin-Madison.